Abstract
Aims/hypothesis
Low mitochondrial activity in adipose tissue is suggested to be an underlying factor in obesity and its metabolic complications. We aimed to find out whether mitochondrial measures are downregulated in obesity also in isolated adipocytes.
Methods
We studied young adult monozygotic (MZ) twin pairs discordant (n = 14, intrapair difference ΔBMI ≥ 3 kg/m2) and concordant (n = 5, ΔBMI < 3 kg/m2) for BMI, identified from ten birth cohorts of 22- to 36-year-old Finnish twins. Abdominal body fat distribution (MRI), liver fat content (magnetic resonance spectroscopy), insulin sensitivity (OGTT), high-sensitivity C-reactive protein, serum lipids and adipokines were measured. Subcutaneous abdominal adipose tissue biopsies were obtained to analyse the transcriptomics patterns of the isolated adipocytes as well as of the whole adipose tissue. Mitochondrial DNA transcript levels in adipocytes were measured by quantitative real-time PCR. Western blots of oxidative phosphorylation (OXPHOS) protein levels in adipocytes were performed in obese and lean unrelated individuals.
Results
The heavier (BMI 29.9 ± 1.0 kg/m2) co-twins of the discordant twin pairs had more subcutaneous, intra-abdominal and liver fat and were more insulin resistant (p < 0.01 for all measures) than the lighter (24.1 ± 0.9 kg/m2) co-twins. Altogether, 2538 genes in adipocytes and 2135 in adipose tissue were significantly differentially expressed (nominal p < 0.05) between the co-twins. Pathway analysis of these transcripts in both isolated adipocytes and adipose tissue revealed that the heavier co-twins displayed reduced expression of genes relating to mitochondrial pathways, a result that was replicated when analysing the pathways behind the most consistently downregulated genes in the heavier co-twins (in at least 12 out of 14 pairs). Consistently upregulated genes in adipocytes were related to inflammation. We confirmed that mitochondrial DNA transcript levels (12S RNA, 16S RNA, COX1, ND5, CYTB), expression of mitochondrial ribosomal protein transcripts and a major mitochondrial regulator PGC-1α (also known as PPARGC1A) were reduced in the heavier co-twins’ adipocytes (p < 0.05). OXPHOS protein levels of complexes I and III in adipocytes were lower in obese than in lean individuals.
Conclusions/interpretation
Subcutaneous abdominal adipocytes in obesity show global expressional downregulation of oxidative pathways, mitochondrial transcripts and OXPHOS protein levels and upregulation of inflammatory pathways.
Data availability
The datasets analysed and generated during the current study are available in the figshare repository, https://dx.doi.org/10.6084/m9.figshare.3806286.v1
Similar content being viewed by others
Introduction
Adipose tissue is the most flexible organ of the body, readily adapting to changes in energy supply. Mitochondria are essential in these adaptations, as their molecular machinery governs the metabolic pathways by which nutrients are either oxidised or stored [1]. The mitochondria’s own genome [2] and over 1500 genes from the nucleus [3] control the function and the energy adaptations of the mitochondria. Low mitochondrial biogenesis has previously been suggested to characterise adipose tissue in morbid obesity [4], insulin resistance [5] and type 2 diabetes [6], although evidence of this in healthy obesity has been lacking. In our recent study, we demonstrated that mitochondria-related transcripts and pathways, mitochondrial DNA (mtDNA) amount, mtDNA-encoded transcripts and oxidative phosphorylation (OXPHOS) protein levels are reduced in adipose tissue of obese vs lean monozygotic (MZ) co-twins [7]. In addition, mitochondrial content measured by mtDNA copy-number [8, 9] and OXPHOS transcripts [9, 10] have been shown to be downregulated in adipose tissue in obesity, and basal oxygen consumption is lower in adipose tissue of obese compared with lean individuals [11].
However, since adipose tissue is a mixture of adipocytes and stromal vascular fraction cells (SVFCs), the sole contribution of adipocytes to the dysfunction of mitochondria in the tissue is not yet known. While comparative data for adipocytes and whole adipose tissue are lacking, a few previous studies have examined mitochondrial variables in either mature adipocytes [12, 13] or pre-adipocytes [14] in obesity. Pre-adipocytes differentiated in vitro from human subcutaneous adipose tissue (SAT) of obese persons had lower oxygen consumption rates after β-adrenergic stimulation compared with that from lean individuals [14]. A recent study suggests that both mitochondrial oxygen consumption rates and mitochondrial content are reduced in adipocytes in obesity, independent of adipocyte size [13]. Another study proposes that the reduced oxidative capacity of adipocyte mitochondria in obesity is related to overall adiposity rather than cell size [12]. However, based on data from unrelated obese and lean groups, the genetic and acquired effects on mitochondrial function cannot be distinguished. Further, the mitochondrial transcriptomics pathways and their relationships with whole-body metabolism have not been studied before in adipocytes in obesity.
We set out to study the effects of acquired obesity on transcriptomics profiles of adipocytes, with a specific focus on mitochondria, in a rare sample of MZ twin pairs discordant for BMI. This design controls for genes, sex, age and early environmental factors between obese and lean individuals by comparing heavier and lighter co-twins within BMI-discordant pairs. In addition to nuclear transcriptomics pathways targeting mitochondria in both adipocytes and adipose tissue, we measured transcript levels of genes encoded by mtDNA and OXPHOS protein levels in adipocytes.
Methods
Participants
The participants were from two population-based birth cohorts of Finnish twins: 1975–1979 (FinnTwin16, n = 2839 pairs) and 1983–1987 (FinnTwin12, n = 2578 pairs), aged 23.5–35.8 years at the time of the study [15]. Altogether, of the 26 MZ twin pairs who were discordant for BMI (within-pair difference, ΔBMI ≥ 3 kg/m2, and reported previously [7, 16]); 14 healthy pairs (three men, 11 women, aged 28.5 ± 0.9 years) had adipocytes available for the present study. Five healthy MZ twin pairs concordant for BMI (ΔBMI < 3 kg/m2, one man, four women, aged 29.7 ± 1.0 years) were included as controls. The mtDNA-encoded transcript levels were determined from 12 discordant twin pairs. The twins had no diseases and took no regular medications except for contraceptives. In addition, six obese (mean BMI 40 ± 2.2 kg/m2; one man, five women, aged 49 ± 2.8 years, three on hypertensive medication) and seven lean (mean BMI 19.9 ± 0.3 kg/m2; one man, six women, aged 31 ± 1.5 years) individuals provided adipocytes for western blot analyses (see electronic supplementary material [ESM] Methods). Written informed consent was obtained from all participants. The study protocol was designed and performed according to the principles of the Helsinki Declaration and approved by the Ethical Committee of the Helsinki University Central Hospital.
Clinical assessments and analytical measurements
Height and weight were measured, after participants had fasted overnight, to calculate BMI. Body composition (dual-energy x-ray absorptiometry) [17], volumes of SAT and visceral adipose tissue (VAT) (MRI), liver fat content (magnetic resonance [MR] spectroscopy) [18] and plasma glucose and insulin for calculation of HOMA and Matsuda indices, serum high-sensitivity C-reactive protein (hs-CRP), plasma leptin, adiponectin and physical activity were measured (see ESM Methods), as previously described [19].
Adipose tissue biopsies, adipocyte extraction and RNA extraction
Surgical biopsy samples of abdominal SAT were collected under local anaesthesia and snap-frozen in −80°C liquid nitrogen. Adipocytes were collected by digesting a piece of fresh adipose tissue in 2% collagenase–DMEM/F-12 (see ESM Methods). Total adipose tissue and adipocyte RNA was extracted using RNeasy Lipid Tissue Mini Kit (Qiagen, Nordic, Solletuna, Sweden) with a DNase I (Qiagen) digestion according to the manufacturer’s instructions.
Part of the collagenase-digested adipose tissue sample was used for measurement and calculation of adipocyte volume and number (see ESM Methods) [19].
Transcriptomics and pathway analyses in adipocytes and adipose tissue
Total RNA (500 ng) from adipocytes and adipose tissue was used for gene expression analysis on an Affymetrix U133 Plus 2.0 array (Thermo Fischer, Santa Clara, CA, USA) according to the manufacturer’s instructions. Pre-processing of the expression data was done with BioConductor software (open source, www.bioconductor.org), the GC-RMA algorithm and annotation by Brainarray CDF version 18 [20]. Differential expression between the co-twins was analysed with Limma (a package of Bioconductor, open source) [21], thereafter subjecting the significant genes to Qiagen’s Ingenuity Pathway Analysis (IPA; Qiagen Redwood City, www.qiagen.com/ingenuity, accessed 1 August 2015). The top ten significant pathways were chosen for further examination and calculation of the mean centroid values (indicating mean pathway activation of genes listed in Table 4) [9]. Mean centroids were also calculated for the expression of small (MRPS, 29 transcripts in Affymetrix) and large (MRPL, 49 transcripts) mitochondrial ribosomal proteins (see our additional data on figshare https://dx.doi.org/10.6084/m9.figshare.3806286.v1 data S1) [22]). MitoCarta [3], an online atlas of 1013 nuclear and mtDNA genes encoding proteins of mitochondrial localisation, was used to select mitochondria-related transcripts among the significantly differentially expressed genes between the co-twins.
Additionally, the significantly differentially expressed genes between the co-twins that were consistently up- or downregulated in the heavier co-twin, in at least 12 out of the 14 discordant twin pairs, were analysed with IPA pathway analysis.
qRT-PCR of mtDNA-encoded transcripts and PGC-1α in adipocytes
A total of 300 ng of RNA from adipocytes, synthesised to complementary DNA (cDNA) (iScript cDNA Synthesis Kit, 170-8891; Bio-Rad, Hemel Hempstead UK), was used in the quantitative reverse transcription PCR (qRT-PCR) analyses (Bio-Rad) of mtDNA-encoded 12S rRNA (MT-RNR1), 16S rRNA (MT-RNR2), mRNAs (MT-COX1, MT-ND5, MT-CYTB and PGC-1α [also known as PPARGC1A]) (IQ Custom SYBR Green Supermix 170-8860; Bio-Rad) [7] (see ESM Methods). The nuclear-encoded 18S rRNA, GAPDH, YWHAZ and IPO8 genes were used as controls. The primer sequences are shown in ESM Table 1. The data were analysed by qBASE+ (version 2.6.1, Biogazelle, Zwijnaarde, Belgium) software.
Western blot for OXPHOS protein levels in adipocytes
For the western blot, 10 μg of adipocyte protein lysates (obtained by homogenisation in RIPA buffer) from six obese and seven lean individuals was separated on 4–20% SDS-PAGE gels. The primary antibodies were Total OXPHOS antibody mixture (110411; Abcam, Cambridge, UK); NDUFB8 (complex I), SDHB (complex II), UQCRC2 (complex III), COX2 (complex IV) and ATP5A (complex V). Additionally, individual antibodies were used for complex III core 2 subunit (MS304) and complex V subunit α (MS507). Actin (Santa Cruz Biotechnology, Heidelberg, Germany), vinculin (Abcam ab129002) and porin (Abcam) were used as controls. Total OXPHOS and vinculin antibodies were incubated in 5% (wt/vol.) milk/Tris-buffered saline (154 mmol/l NaCl) and 0.1% (vol./vol.) Tween 20 (TBST) , and complex III-core 2 subunit, complex V subunit α, actin and porin antibodies in 1% (wt/vol.) BSA/TBST at concentrations of 1:1000 (vinculin at 1:10,000) overnight at 4°C. Incubation with secondary horseradish peroxidase-conjugated anti-mouse (Santa Cruz), anti-rabbit (Jackson ImmunoResearch, Baltimore, NJ, USA) and anti-goat (401504; Calbiochem, Darmstadt, Germany) (1:2500) was carried out in 5% (wt/vol,) milk–TBST for 1 h at room temperature (see ESM Methods). The proteins were quantified with ImageLab v3.0 software (Bio-Rad, Hercules, CA, USA).
Statistical analyses
Statistical analyses were performed using Stata statistical software (Release 13.0; Stata Corporation, College Station, TX, USA). Differences between the co-twins were calculated by Wilcoxon’s signed rank tests and differences in the protein levels between lean and obese individuals by Mann–Whitney U test. Correlations between pathways and anthropometric and metabolic measures in all twin individuals were calculated using Pearson correlations, corrected for clustered sampling of co-twins by survey methods [23]. Non-normally distributed variables (VAT, liver fat, adipocyte volume, HOMA index, Matsuda, leptin, adiponectin and hs-CRP) were logarithmically (base 10) corrected for analyses. Adjustments for multiple comparisons in Table 5 were made by the number of principal components analysis (PCA) to provide a proper p value threshold. The PCA correction for the mitochondrial variables produced two principal component factors, which explained 78% of the variance. Therefore, the adjusted significance level for analyses in Table 5 was p < 0.05 divided by two (i.e. p < 0.025). For ESM Table 3 (adipose tissue) the analysis provided one principal component explaining 81% of the variance, and an adjusted p value of 0.05.
Results
The heavier (mean ± SE BMI 29.9 ± 1.0 kg/m2) and lighter (24.1 ± 0.9 kg/m2) co-twins of the discordant pairs had a mean difference in body weight of 16.9 ± 1.8 kg (p < 0.001) (Table 1). The heavier co-twins had more SAT, VAT and liver fat, larger adipocytes and were more insulin resistant (p < 0.05 all, Table 1). No significant differences in fat or metabolic measures were found between the concordant co-twins, despite a small difference in BMI (light 28.2 ± 1.9 vs heavy 30.4 ± 1.8 kg/m2, p = 0.043, Table 1).
Over 2000 genes were differentially expressed in adipocytes within the discordant pairs, revealing mitochondrial associations in pathway analyses
We first analysed the genome-wide differences in adipocytes of the lighter vs heavier discordant co-twins. A total of 2538 transcripts (figshare data S2 [22]) were differentially expressed within the twin pairs (nominal p value < 0.05). The top ten up- and downregulated genes are presented according to p value in Table 2 and fold change (FC) in Table 3, and figshare data S2 [22].
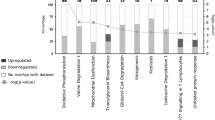
The top ten pathways of the significantly differentially expressed genes between the co-twins in adipocytes from IPA analyses (all p < 0.001) were subjected to calculation of the mean centroid values, representing the global transcriptional activity of the pathway. Based on the mean centroids, the significantly downregulated pathways in adipocytes of the heavier co-twins were OXPHOS, glutaryl-CoA degradation, mTOR signalling and branched-chain amino acid (BCAA) catabolism (valine and isoleucine degradation) and the significantly upregulated pathways were glucocorticoid receptor and IL-8 signalling (p < 0.05 all, Wilcoxon’s signed rank test, Table 4 and figshare data S3 [22]). Mitochondria-related pathways of OXPHOS and valine, isoleucine and glutaryl-CoA degradation correlated negatively with many adiposity and insulin resistance measures and hs-CRP, and positively with Matsuda index and adiponectin (Table 5). For IL-8 receptor signalling and glucocorticoid receptor signalling, the correlations were opposite to those seen for the above mentioned mitochondrial pathways.
Upstream regulator analyses and reduced levels of a major mitochondrial regulator PGC-1α in adipocytes
To detect transcripts regulating the observed genes and pathways, we performed an IPA upstream regulator analysis for the significantly differentially expressed genes in adipocytes. The top three significant regulators according to IPA were SREBF1, CEBPA and MYC (Table 6 and figshare S4 [22]). However, in the subsequent Wilcoxon’s test of these transcripts, none were significantly different between the co-twins.
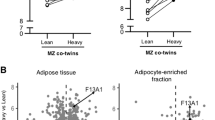
When using the differentially expressed genes combined with the MitoCarta gene list (figshare data S5 [22]) to detect specifically mitochondria-related regulators, SREBF1, MYC and PGC-1α (Table 6 and figshare data S6 [22]) emerged as the top three regulators. PGC-1α was the only transcription factor significantly reduced in the heavier compared with the lighter co-twins (3.9 ± 0.3 vs 4.3 ± 0.3 Affymetrix units, respectively, p = 0.0157, Fig. 1a). This result was confirmed by qRT-PCR (heavier 2.6 ± 0.6 vs lighter 4.1 ± 1.0, p = 0.0414, Fig. 1b).
Reduced levels of mitochondrial regulator, PGC-1α, in adipocytes of the heavier co-twins. (a) Affymetrix expression. (b) qRT-PCR, n=14 discordant co-twins; 18S RNA, hGAPDH, YWHAZ and IPO8 as control genes. White bars, lighter twins; black bars, heavier twins. Wilcoxon’s rank sum test, *p < 0.05 vs the lighter twins. Data is presented as mean ± SEM
Consistently up- and downregulated transcripts in adipocytes revealed mitochondria-related transcriptional downregulation and immune-related upregulation
Next, we focused on the transcripts that were not only significantly different between the co-twins but also consistently up- or downregulated in the heavier co-twins, in at least 12 out of 14 discordant twin pairs. Adipocytes had 454 consistently upregulated and 538 consistently downregulated transcripts in the heavier co-twins (figshare data S7 [22]). The downregulated genes revealed mitochondria-related pathways in IPA (OXPHOS, fatty acid β-oxidation, AMP-activated protein kinase signalling, glutaryl-CoA degradation, tricarboxylic acid [TCA] cycle; p < 0.001 all), while the upregulated pathways displayed an immune-related pattern (IL-10 signalling, granulocyte adhesion, IL-8 signalling, recognition of bacteria and viruses, high mobility group box 1 (HMGB1) signalling and cytokine production; p < 0.001 all, figshare data S8 [22]).
Reduction of mtDNA transcript levels (qRT-PCR) and mitochondrial ribosomal protein subunit gene expression in the heavier co-twins’ adipocytes
The above results strongly suggest that genes targeting mitochondria are downregulated in the heavier co-twins’ adipocytes. Because Affymetrix only captures nuclear genes, we next analysed the expression of genes encoded by mtDNA by qRT-PCR. Here, the mitochondrial rRNAs (12S, MT-RNR1, p = 0.0022 and 16S, MT-RNR2, p = 0.0186) and mRNAs (MT-COX1, CIV subunit, p = 0.0047, MT-ND5, CI subunit, p = 0.0186, and MT-CYTB, CIII subunit, p = 0.0047) were significantly downregulated in the heavier as compared with the lighter co-twins (Fig. 2).
Reduced levels of mitochondrial mtDNA transcripts (qRT-PCR; rRNA’s 12S RNA, 16S RNA and mRNA’s MT-COX1, MT-ND5, MT-CYTB) in the heavier co-twins’ adipocytes (n=12 discordant co-twins); 18S RNA, hGAPDH, YWHAZ and IPO8 as control genes. White bars, lighter twins; black bars, heavier twins. Wilcoxon’s rank sum test, *p < 0.05 and **p < 0.01 vs lighter twins. Data is presented as mean ± SEM
The mitochondrial rRNAs and mRNAs are translated into proteins by mitochondrial ribosomes. The mean centroids of both small (−0.24 ± 0.18 vs 0.23 ± 0.18 arbitrary Affymetrix units [AU], p = 0.0092) and large (−0.17 ± 0.18 vs 0.18 ± 0.17 AU, p = 0.0157) mitochondrial ribosomal protein subunit transcripts were downregulated in the heavier compared with the lighter co-twins’ adipocytes (Fig. 3 and figshare data S1 [22]).
Reduction of OXPHOS protein levels in adipocytes of obese vs lean individuals
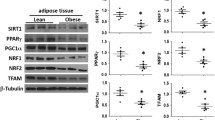
The mitochondrial OXPHOS protein CI subunit NDUFB8 (p = 0.0176) and CIII-core 2 subunit (CIII, p = 0.0455) were significantly reduced in the obese compared with the lean individuals, whereas COX2 (CIV subunit, p = 0.253), ATP5A (CV subunit, p = 0.254) and the nuclear-encoded CII subunit SDHB (p = 0.361) were not (Fig. 4). When the OXPHOS protein signals were normalised against the mitochondrial protein porin, complex CI subunit NDUFB8 (p = 0.0106) was significantly reduced in the obese compared with the lean individuals, suggesting a possible decrease in OXPHOS CI levels per mitochondria. CIV levels decreased non-significantly (p = 0.088) and CII (p = 0.988), CIII (p = 0.199) and CV (p = 0.775) levels were unchanged (Fig. 4).
Reduced levels of OXPHOS protein subunits CI–CV (a) per cell (compared with cytosolic actin) and (b) per mitochondria (compared with mitochondrial porin) in the obese (n=6, black bars) compared with the lean (n=7, white bars) individuals. Wilcoxon’s rank sum test, *p < 0.05, † p = 0.08 vs lean individuals. Data is presented as mean ± SEM
Adipocyte volume and number
Whether adipocyte size matters in reduced mitochondrial biogenesis or the metabolic complications in obesity is debatable, so we analysed the associations of adipocyte size and number with the above gene expression results. Adipocyte volume was significantly larger in the heavier than in the lighter co-twins, while the number of adipocytes did not differ between the co-twins (Table 1). Adipocyte volume correlated negatively with mitochondrial glutaryl-CoA degradation and mTOR signalling and positively with inflammatory IL-8 signalling for adipocytes (Table 5). However, in a multiple regression analysis adjusting for body fat mass (kg) and sex, adipocyte volume did not yield independent associations with the gene expression pathways (data not shown).
Adipose tissue transcriptomic analyses mimicked those of the adipocytes
To compare the gene expression results from adipocytes with those from adipose tissue, we performed the same transcriptional analyses of the 14 discordant co-twins in adipose tissue. A total of 2135 transcripts (figshare data S2 [22]) were differentially expressed between the co-twins (nominal p value < 0.05). As in adipocytes, the top ten upregulated genes in the heavier co-twins were associated with immune reactions and the top ten downregulated transcripts (figshare data S2 [22]) were associated with mitochondria, fatty acid synthesis and β-oxidation. Analyses according to the largest FC between the co-twins revealed chemokines and inflammatory genes among the top upregulated list, as in adipocytes. Downregulated transcripts included mostly the same genes as in adipocytes.
The pathway analyses in adipose tissue mimicked those of adipocytes, with OXPHOS, valine and lysine degradation, glutaryl-CoA degradation, acetate conversion to acetyl-CoA, fatty acid β-oxidation, triacylglycerol synthesis and ketone body production and breakdown among the top ten pathways; the mean centroids of these were all downregulated in the heavier co-twins (p < 0.05 all, Wilcoxon’s signed rank test, ESM Table 2 and figshare data S3 [22]). In general, significant negative correlations were found for the mean centroids of the top ten pathways and adiposity, including adipocyte size, leptin, insulin resistance and hs-CRP and positive correlations with Matsuda index and adiponectin (ESM Table 3). Triacylglycerol synthesis correlated only with VAT and liver fat.
The top three upstream regulators in adipose tissue were BACH1, CEBPA and ERG, the mean centroids of which did not differ between the co-twins. When combining the gene list with MitoCarta (figshare data S5 [22]) to analyse the mitochondrial upstream regulators in adipose tissue, SREBF1, PGC-1α and EIF2A (figshare data S6 [22]) emerged as the top three regulators. PGC-1α (3.8 ± 0.3 vs 4.9 ± 0.3 AU, p = 0.0019 ESM Fig. 1a) and EIF2A (11.2 ± 0.04 vs 11.4 ± 0.1 AU, p = 0.0029) were downregulated in the heavier co-twins.
In consistency analyses we found 221 upregulated and 437 downregulated transcripts (figshare data S7 [22]) in the heavier co-twins’ adipose tissue. The top significantly downregulated pathways included, for example, cell cycle control, DNA modifications, mTOR signalling and cholesterol biosynthesis, and upregulated pathways mostly immune- and lipid-related signalling (p < 0.001 all, IPA, figshare data S8 [22]).
In our previous study with 26 BMI-discordant pairs [7], we reported reduced expression of mitochondrial ribosomal protein subunit transcripts in the heavier co-twins’ adipose tissue. In the current study, we repeated these ribosomal protein subunit expression level analyses in adipose tissue in the 14 discordant pairs. Both MRPS (heavier twins −0.24 ± 0.19 vs lighter twins 0.11 ± 0.14 AU, p < 0.01) and MRPL (heavier twins −0.21 ± 0.2 vs lighter twins 0.01 ± 0.15 AU, p < 0.05, figshare data S1 [22]) were downregulated in the heavier co-twins (ESM Fig. 1b), as in adipocytes.
Discussion
This study of BMI-discordant MZ twin pairs shows that mitochondria-related transcriptional signature, at the nuclear and mitochondrial transcription level, is downregulated in adipocytes in the heavier co-twins. This was particularly the case in the consistency analyses: the genes that were consistently downregulated in at least 12 out of the 14 heavier co-twins were involved in mitochondrial pathways. Subsequently, expression results of PGC-1α, the major regulator of mitochondrial biogenesis, the mtDNA-encoded genes and the mitochondrial ribosomes as well as reduced levels of OXPHOS CI and CIII subunits all demonstrated that mitochondrial oxidative metabolism is downregulated in adipocytes of individuals with higher body weight. Thus, we suggest that adipocytes are major contributors to the downregulation of mitochondrial oxidative pathways and reduction in OXPHOS protein levels in obese adipose tissue. The most consistently upregulated genes in the heavier co-twins’ adipocytes clustered to inflammatory pathways suggesting that inflammatory signature arises also from purified adipocytes. Simultaneous suppression of mitochondria and activation of inflammatory gene expression suggests that these two phenomena may be biologically linked.
Our study reports similarities in the gene expression patterns of subcutaneous adipocytes and SAT in the BMI-discordant twin pairs. Notably, genes clustering to mitochondrial pathways were the most significantly downregulated in the heavier co-twins both in adipocytes and in adipose tissue. We have previously demonstrated a widespread downregulation of mitochondria- and nuclear-encoded mitochondria-related genes, reduction of mtDNA amount and reduction of OXPHOS protein subunit levels in adipose tissue of heavier vs leaner co-twins [7]—alterations that were associated with fatty liver, insulin resistance and low-grade inflammation [7, 16]. We have also shown reduced expression of mitochondrial pathways and upregulated inflammatory pathways in adipose tissue in obesity [9]. Other studies have also reported downregulation of mitochondrial oxidative metabolism in obesity. Oxidative capacity was lower in adipose tissue of obese mice compared with lean mice [24, 25] and in humans, membrane potential, the activity of the respiratory chain complexes I–IV and phosphate utilisation were reduced in isolated mitochondria from obese SAT [26]. The studies on adipose tissue, however, include SVFCs that contribute to the results. In recent studies in adipocytes, the oxygen consumption rates [12] and subunit levels of complexes I and IV [13] were lower in obese vs lean individuals. In contrast, Dahlman et al suggested that the downregulation of electron transport chain genes in VAT of women with type 2 diabetes would be independent of obesity [27]. In our study we had the opportunity to compare gene expression of subcutaneous adipocytes and SAT in MZ discordant co-twins and examine the effects of acquired excess weight. Our results show that both adipocytes and adipose tissue are remarkably similar regarding the expression and downregulation of mitochondria-related genes and pathways in the heavier co-twins, suggesting that adipocytes seem to be a major contributor to the reduced mitochondrial oxidative metabolism of adipose tissue in acquired obesity.
Strongly upregulated inflammatory gene expression was found in adipocytes and adipose tissue of the heavier co-twins. Adipose tissue of obese persons is characterised by accumulation of macrophages and other immune cells [28, 29]. Inflammatory cells release inflammatory cytokines in obese adipose tissue [28, 30, 31]. However, adipocytes are also known secretors of IL-6, IL-8, TNF-α and inflammatory adipokines [32], and exhibit immune-cell-like functions that trigger CD4+ T cell inflammation, independent of macrophages [33]. Macrophage-derived TNF-α enhances the expression of inflammatory genes in murine adipocytes [34, 35]. In humans, adipose tissue TNF-α expression and secretion is increased in obesity [36, 37] and may mediate insulin resistance [36, 38]. However not all studies agree [37, 39]. Adipocytes also express high levels of CD14 and CD68 [40], previously considered as being macrophage-specific. The upregulation of inflammatory genes and pathways in the heavier co-twins in our study supports findings that adipocytes do contribute to the amplified inflammatory milieu of obese adipose tissue, even though inflammation in the whole adipose tissue most probably arises also from the SVFCs.
In our study, we report the associations of mitochondrial downregulation in adipocytes and adipose tissue with increased adiposity (subcutaneous, visceral, liver fat) and impaired metabolism (reduced insulin sensitivity, plasma adiponectin and increased hs-CRP levels). However, the direction of causality between mitochondrial downregulation and metabolic impairments is not clear. In fat-specific Tfam-knockout mice, increased mitochondrial oxidation protected from the development of diet-induced obesity, hepatosteatosis and insulin resistance [41]. In contrast, studies in high-fat-fed mice have suggested that reduction in adipose tissue mitochondrial content occurs after the development of impaired glucose homeostasis [42, 43]. In a murine model of obesity, however, mitochondrial respiratory capacity in adipocytes was reduced irrespective of the glucose tolerance status [44]. In humans, genes related to mitochondrial oxidative metabolism at the transcription level have been shown to be downregulated in type 2 diabetes and in the VAT but not SAT depots [27].
In our study, adipocyte volume had significant negative correlations with mitochondrial pathways in adipocytes and adipose tissue. Enlargement of adipocytes is a main feature in obesity [45, 46]. Failure of adipocyte differentiation and pre-adipocyte recruitment is thought to result in excess adipocyte hypertrophy and in insulin-resistant adipocytes [47, 48]. This could lead to metabolic problems [19] and could be a means of distinguishing between metabolically unhealthy and healthy obese individuals [16]. However, after adjusting for body fat mass (kg) and sex, mitochondrial pathways and adipocyte size were not correlated in our study, indicating that reduction in mitochondrial transcription was due to general fat accumulation rather than cell size. Some recent studies also supported this view by showing that adipocyte mitochondrial respiration is reduced in human obesity independent of adipocyte size [12, 13] and demonstrated that the compromised respiratory capacity may be a general feature of all adipocytes from an obese individual, not only of the enlarged adipocytes [13].
Studies in unrelated individuals raise the question of whether adipocytes from obese individuals are genetically equipped with poor mitochondria or whether this is an acquired phenomenon. The MZ co-twin setting gives a unique opportunity to control for genetic and early environmental factors. In our data, the reduced mitochondria-related transcription in obese adipocytes is clearly acquired. However, because our study is cross-sectional, we cannot make definitive cause-and-effect conclusions. Since the BMI discordance in our twins only became evident in young adulthood [7, 49], it is likely that the adipocyte mitochondria within the twin pairs were similar until that period of life. Because we were not able to measure the mitochondrial pathways of SVFCs, we cannot make conclusions on how strong the influence of adipocytes is on mitochondria-related transcriptional downregulation in relation to SVF cells of adipose tissue. In the absence of the measures of mitochondrial function, mRNA measures alone do not provide direct evidence of the physiological dysfunction of mitochondria. The low level of significantly different transcripts, corrected for false-discover rate, between the co-twins in the adipocyte transcriptomics data may be related to the small number of twin pairs in the study. The studied twin pairs were also healthier than the other 12 twin pairs included in our previous analyses [7]. Although we saw clear trends for lower protein levels in the obese individuals, in most cases the statistical power was too low to detect significant p values. The lean and obese unrelated individuals had larger differences related to age and BMI compared with the twins. Due to limited sample material, the mtDNA amount could not be measured. Because of the small sample size and the associative nature of the study, the results may need to be confirmed with larger data.
In this study we demonstrate for the first time that gene expression differences in acquired obesity are similar in adipocytes and adipose tissue of young MZ BMI-discordant co-twins. Mitochondria-related nuclear pathways and transcripts were downregulated in adipocytes and adipose tissue, and mtDNA transcript levels and levels of CI and CIII OXPHOS subunits reduced in adipocytes, from individuals with larger body weight. Our results suggest that downregulation of the essential functions of mitochondria, such as OXPHOS, BCAA catabolism, TCA cycle, fatty acid β-oxidation and ketone metabolism, as well as upregulation of inflammation, are key characteristics of adipocytes in obesity.
Abbreviations
- AU:
-
Arbitrary Affymetrix units
- BCAA:
-
Branched-chain amino acids
- cDNA:
-
Complementary DNA
- FC:
-
Fold change
- hs-CRP:
-
High-sensitivity C-reactive protein
- IPA:
-
Ingenuity Pathway Analysis
- MR:
-
Magnetic resonance
- MRPL:
-
Mitochondrial ribosomal protein large subunits
- MRPS:
-
Mitochondrial ribosomal protein small subunits
- mtDNA:
-
Mitochondrial DNA
- MZ:
-
Monozygotic
- OXPHOS:
-
Oxidative phosphorylation
- PCA:
-
Principal components analysis
- qRT-PCR:
-
Quantitative reverse transcription PCR
- SAT:
-
Subcutaneous adipose tissue
- SVFC:
-
Stromal vascular fraction cell
- TBST:
-
Tris-buffered saline–Tween
- TCA:
-
Tricarboxylic acid cycle
- VAT:
-
Visceral adipose tissue
References
De Pauw A, Tejerina S, Raes M, Keijer J, Arnould T (2009) Mitochondrial (dys)function in adipocyte (de)differentiation and systemic metabolic alterations. Am J Pathol 175:927–939
Falkenberg M, Larsson NG, Gustafsson CM (2007) DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem 76:679–699
Pagliarini DJ, Calvo SE, Chang B et al (2008) A mitochondrial protein compendium elucidates complex I disease biology. Cell 134:112–123
Semple RK, Crowley VC, Sewter CP et al (2004) Expression of the thermogenic nuclear hormone receptor coactivator PGC-1α is reduced in the adipose tissue of morbidly obese subjects. Int J Obes Relat Metab Disord 28:176–179
Hammarstedt A, Jansson PA, Wesslau C, Yang X, Smith U (2003) Reduced expression of PGC-1 and insulin-signaling molecules in adipose tissue is associated with insulin resistance. Biochem Biophys Res Commun 301:578–582
Bogacka I, Xie H, Bray GA, Smith SR (2005) Pioglitazone induces mitochondrial biogenesis in human subcutaneous adipose tissue in vivo. Diabetes 54:1392–1399
Heinonen S, Buzkova J, Muniandy M et al (2015) Impaired mitochondrial biogenesis in adipose tissue in acquired obesity. Diabetes 64:3135–3145
Kaaman M, Sparks LM, van Harmelen V et al (2007) Strong association between mitochondrial DNA copy number and lipogenesis in human white adipose tissue. Diabetologia 50:2526–2533
Pietilainen KH, Naukkarinen J, Rissanen A et al (2008) Global transcript profiles of fat in monozygotic twins discordant for BMI: pathways behind acquired obesity. PLoS Med 5:e51
Mustelin L, Silventoinen K, Pietilainen K, Rissanen A, Kaprio J (2009) Physical activity reduces the influence of genetic effects on BMI and waist circumference: a study in young adult twins. Int J Obes (Lond) 33:29–36
Hallgren P, Sjostrom L, Hedlund H, Lundell L, Olbe L (1989) Influence of age, fat cell weight, and obesity on O2 consumption of human adipose tissue. Am J Physiol 256:E467–E474
Yin X, Lanza IR, Swain JM, Sarr MG, Nair KS, Jensen MD (2014) Adipocyte mitochondrial function is reduced in human obesity independent of fat cell size. J Clin Endocrinol Metab 99:E209–E216
Fischer B, Schottl T, Schempp C et al (2015) Inverse relationship between body mass index and mitochondrial oxidative phosphorylation capacity in human subcutaneous adipocytes. Am J Physiol Endocrinol Metab 309:E380–E387
Yehuda-Shnaidman E, Buehrer B, Pi J, Kumar N, Collins S (2010) Acute stimulation of white adipocyte respiration by PKA-induced lipolysis. Diabetes 59:2474–2483
Kaprio J (2006) Twin studies in Finland 2006. Twin Res Hum Genet 9:772–777
Naukkarinen J, Heinonen S, Hakkarainen A et al (2014) Characterising metabolically healthy obesity in weight-discordant monozygotic twins. Diabetologia 57:167–176
Pietrobelli A, Formica C, Wang Z, Heymsfield SB (1996) Dual-energy X-ray absorptiometry body composition model: review of physical concepts. Am J Physiol 271:E941–E951
Graner M, Seppala-Lindroos A, Rissanen A et al (2012) Epicardial fat, cardiac dimensions, and low-grade inflammation in young adult monozygotic twins discordant for obesity. Am J Cardiol 109:1295–1302
Heinonen S, Saarinen L, Naukkarinen J et al (2014) Adipocyte morphology and implications for metabolic derangements in acquired obesity. Int J Obes (Lond) 38:1423–1431
Dai M, Wang P, Boyd AD et al (2005) Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res 33:e175
Gentleman RC, Carey VJ, Bates DM et al (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80
Heinonen S, Muniandy M, Buzkova J et al (2016) Supplementary data for ‘Mitochondria-related transcriptional signature is downregulated in adipocytes in obesity – a study of young healthy MZ twins’. Figshare: https://dx.doi.org/10.6084/m9.figshare.3806286.v1
Rao J, Scott A (1984) On chi-squared tests for multiway contingency tables with cell proportions estimated from survey data. Ann Stat 12:46–60
Wilson-Fritch L, Nicoloro S, Chouinard M et al (2004) Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J Clin Invest 114:1281–1289
Choo HJ, Kim JH, Kwon OB et al (2006) Mitochondria are impaired in the adipocytes of type 2 diabetic mice. Diabetologia 49:784–791
Chattopadhyay M, Guhathakurta I, Behera P et al (2011) Mitochondrial bioenergetics is not impaired in nonobese subjects with type 2 diabetes mellitus. Metabolism 60:1702–1710
Dahlman I, Forsgren M, Sjogren A et al (2006) Downregulation of electron transport chain genes in visceral adipose tissue in type 2 diabetes independent of obesity and possibly involving tumor necrosis factor-α. Diabetes 55:1792–1799
Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AWJ (2003) Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest 112:1796–1808
Cildir G, Akincilar SC, Tergaonkar V (2013) Chronic adipose tissue inflammation: all immune cells on the stage. Trends Mol Med 19:487–500
Fain JN, Madan AK, Hiler ML, Cheema P, Bahouth SW (2004) Comparison of the release of adipokines by adipose tissue, adipose tissue matrix, and adipocytes from visceral and subcutaneous abdominal adipose tissues of obese humans. Endocrinology 145:2273–2282
Fain JN (2006) Release of interleukins and other inflammatory cytokines by human adipose tissue is enhanced in obesity and primarily due to the nonfat cells. Vitam Horm 74:443–477
Lehr S, Hartwig S, Sell H (2012) Adipokines: a treasure trove for the discovery of biomarkers for metabolic disorders. Proteomics Clin Appl 6:91–101
Meijer K, de Vries M, Al-Lahham S et al (2011) Human primary adipocytes exhibit immune cell function: adipocytes prime inflammation independent of macrophages. PLoS ONE 6:e17154
Permana PA, Menge C, Reaven PD (2006) Macrophage-secreted factors induce adipocyte inflammation and insulin resistance. Biochem Biophys Res Commun 341:507–514
Ruan H, Miles PDG, Ladd CM et al (2002) Profiling gene transcription in vivo reveals adipose tissue as an immediate target of tumor necrosis factor-α: implications for insulin resistance. Diabetes 51:3176–3188
Hotamisligil GS, Arner P, Caro JF, Atkinson RL, Spiegelman BM (1995) Increased adipose tissue expression of tumor necrosis factor-alpha in human obesity and insulin resistance. J Clin Invest 95:2409–2415
Kern PA, Saghizadeh M, Ong JM, Bosch RJ, Deem R, Simsolo RB (1995) The expression of tumor necrosis factor in human adipose tissue. Regulation by obesity, weight loss, and relationship to lipoprotein lipase. J Clin Invest 95:2111–2119
Lofgren P, van Harmelen V, Reynisdottir S et al (2000) Secretion of tumor necrosis factor-α shows a strong relationship to insulin-stimulated glucose transport in human adipose tissue. Diabetes 49:688–692
Mohamed-Ali V, Goodrick S, Rawesh A et al (1997) Subcutaneous adipose tissue releases interleukin-6, but not tumor necrosis factor-α, in vivo. J Clin Endocrinol Metab 82:4196–4200
Khazen W, M’bika J, Tomkiewicz C et al (2005) Expression of macrophage-selective markers in human and rodent adipocytes. FEBS Lett 579:5631–5634
Vernochet C, Mourier A, Bezy O et al (2012) Adipose-specific deletion of TFAM increases mitochondrial oxidation and protects mice against obesity and insulin resistance. Cell Metab 16:765–776
Sutherland LN, Capozzi LC, Turchinsky NJ, Bell RC, Wright DC (2008) Time course of high-fat diet-induced reductions in adipose tissue mitochondrial proteins: potential mechanisms and the relationship to glucose intolerance. Am J Physiol Endocrinol Metab 295:E1076–E1083
Wang P, Kuo H, Huang H et al (2014) Biphasic response of mitochondrial biogenesis to oxidative stress in visceral fat of diet-induced obesity mice. Antioxid Redox Signal 20:2572–2588
Schottl T, Kappler L, Fromme T, Klingenspor M (2015) Limited OXPHOS capacity in white adipocytes is a hallmark of obesity in laboratory mice irrespective of the glucose tolerance status. Mol Metab 4:631–642
Bjorntorp P, Gustafson A, Persson B (1971) Adipose tissue fat cell size and number in relation to metabolism in endogenous hypertriglyceridemia. Acta Med Scand 190:363–367
van Harmelen V, Skurk T, Rohrig K et al (2003) Effect of BMI and age on adipose tissue cellularity and differentiation capacity in women. Int J Obes Relat Metab Disord 27:889–895
Virtue S, Vidal-Puig A (2010) Adipose tissue expandability, lipotoxicity and the metabolic syndrome--an allostatic perspective. Biochim Biophys Acta 1801:338–349
Medina-Gomez G, Virtue S, Lelliott C et al (2005) The link between nutritional status and insulin sensitivity is dependent on the adipocyte-specific peroxisome proliferator-activated receptor-γ2 isoform. Diabetes 54:1706–1716
Pietilainen KH, Rissanen A, Laamanen M et al (2004) Growth patterns in young adult monozygotic twin pairs discordant and concordant for obesity. Twin Res 7:421–429
Acknowledgements
We thank the twins for their invaluable contribution to the study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Data availability
The datasets analysed and generated during the current study are available in the figshare repository, https://dx.doi.org/10.6084/m9.figshare.3806286.v1
Funding
The study was supported by Helsinki University Hospital Research Funds (KHP) and grants from the Novo Nordisk Foundation (KHP), Diabetes Research Foundation (SH, KHP), Jalmari and Rauha Ahokas Foundation (KHP), Finnish Foundation for Cardiovascular Research (KHP), Academy of Finland (grants 265240, 263278 to JK, 272376 and 266286 to KHP), Orion Foundation (SH), Emil Aaltonen Foundation (SH), Maud Kuistila Foundation (SH), Finnish Medical Foundation (SH), Paulo Foundation (SH), Biomedicum Helsinki Foundation (SH), Knut and Alice Wallenberg Foundation (AM), University of Helsinki (KHP), Fondo de Investigación Sanitaria-FEDER PI12/00515 (GF) and PI13/01430 (AR) and CIBEROBN (GF) from the Instituto de Salud Carlos III, Spain.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
SH performed and designed the research, analysed the data, performed the laboratory work and wrote the manuscript. KHP and ARi designed the research, supervised the work and participated in discussion and revision of the results. JK was responsible for selecting the population cohorts from which the discordant pairs were acquired for the study. KHP and SH collected the data. MM performed the whole-genome transcriptomics data analysis. AM analysed the consistently up-and downregulated genes and pathways. AH, JL and NL measured and analysed the MRI and MR spectroscopy data. JB participated in the protein analyses. ARo and GF designed the protocol and participated in the acquisition of the isolate adipocytes. All authors participated in the revision of the manuscript and approved the final version to be published. KHP is the guarantor of this work and as such had full access to the data and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM
(PDF 350 kb)
Rights and permissions
About this article
Cite this article
Heinonen, S., Muniandy, M., Buzkova, J. et al. Mitochondria-related transcriptional signature is downregulated in adipocytes in obesity: a study of young healthy MZ twins. Diabetologia 60, 169–181 (2017). https://doi.org/10.1007/s00125-016-4121-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-016-4121-2